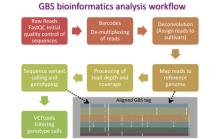
The James Hutton Institute played an active role in the sequencing of the potato genome (The Potato Genome Sequencing Consortium 2011) and the construction of reference potato pseudomolecules (Sharma et al., 2013). Building on these efforts we have established modern sequence-based genotyping platforms for potato genetics including genotyping-by-sequencing (GBS) and whole-exome capture (WEC) approaches. GBS provides a non-targeted but simple, low-cost and highly flexible high-throughput genotyping platform which is being deployed for a range of genetic studies. Genome-wide association study performed at Hutton using GBS has yielded a vast number (~220,000) of robust and well annotated potato SNPs (Sharma et al., unpublished), a subset of which has also been included in the latest version of the Illumina Infinium SNP genotyping array.
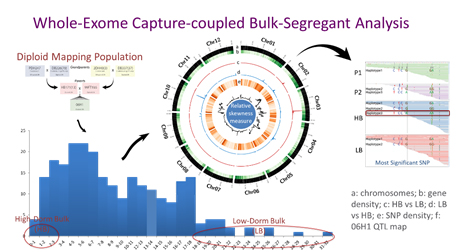 We are also using GBS for characterizing breeding populations and larger germplasm collections where the latter, under the EU-H2020 G2PSOL project, involves genotyping of ~10k potato lines. WEC is a targeted and low-throughput genotyping approach and our design (Sharma et al., unpublished) includes all 39k potato genes as well as reported resistance genes and microRNA targets. We have applied WEC for performing bulk-segregant analysis (BSA) targeting important traits such as tuber dormancy/sprouting and resistance to tobacco-rattle virus. WEC-coupled-BSA approach has proved very effective in revealing QTL regions with high resolution enabling precision in identifying candidate genes and underlying variants for further downstream applications.
We are also using GBS for characterizing breeding populations and larger germplasm collections where the latter, under the EU-H2020 G2PSOL project, involves genotyping of ~10k potato lines. WEC is a targeted and low-throughput genotyping approach and our design (Sharma et al., unpublished) includes all 39k potato genes as well as reported resistance genes and microRNA targets. We have applied WEC for performing bulk-segregant analysis (BSA) targeting important traits such as tuber dormancy/sprouting and resistance to tobacco-rattle virus. WEC-coupled-BSA approach has proved very effective in revealing QTL regions with high resolution enabling precision in identifying candidate genes and underlying variants for further downstream applications.


