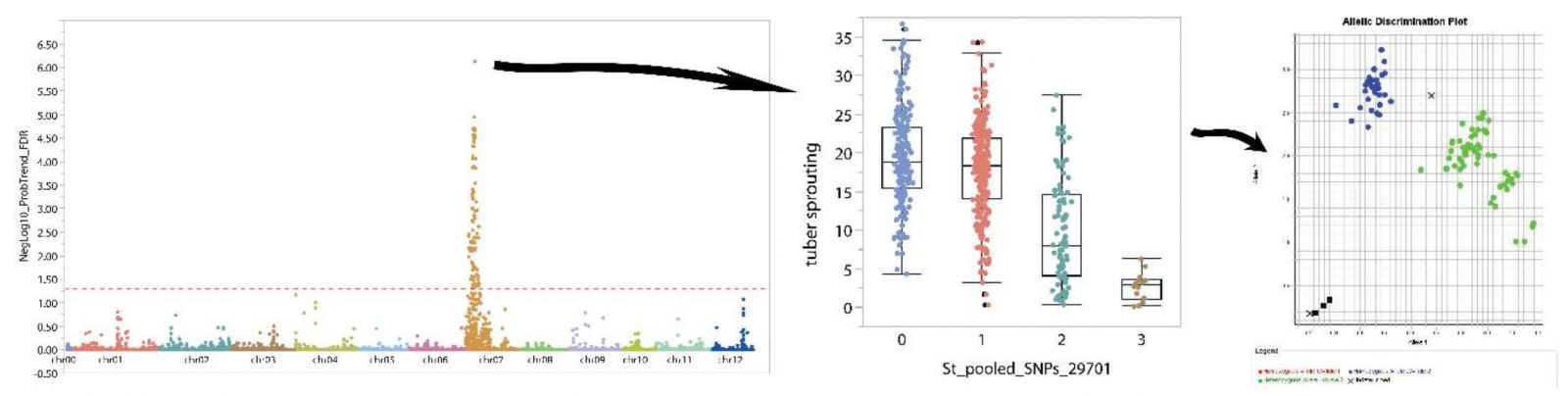
Genome-wide association studies using sequence-based genotyping

The James Hutton Institute has established a diverse and genetically well-characterized association panel (POTAP) of ~350 tetraploid potato lines which we are exploiting to perform genetic studies for various important traits. Genome-wide association studies performed on this panel as part of the TSB (now Innovate UK) Genomes UK grant and in collaboration with our Industrial partners (Cygnet PB Ltd, McCain Potato and James Hutton Ltd) and the AHDB revealed many associations for more than 20 important potato traits (Sharma et al., 2018). POTAP has been extensively genotyped using a ‘fixed’ genotyping platform (8k Illumina Infinium SNP array) as well as flexible genotyping approaches where the latter involves de novo SNP discovery using genotyping-by-sequencing (GBS) approach adapted in-house for potato by researchers at The Hutton (Sharma et al., unpublished).
POTAP has been phenotyped for several important traits yielding a vast number of well-annotated SNPs and marker-trait associations generating a ready platform for performing genomics-assisted breeding and developing diagnostic markers for target traits as well as initiating genomic selection in potato.